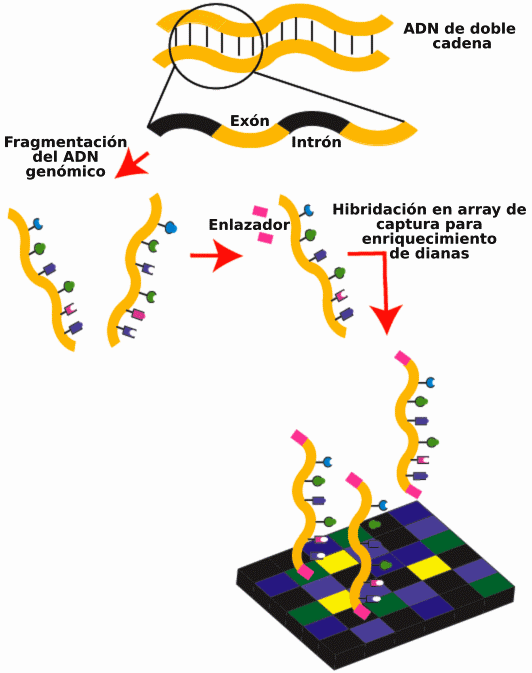
Targeted sequencing, also known as capture sequencing, is a method used to selectively sequence specific regions of DNA or RNA. This technique enables researchers to study only the areas of interest in a sample, rather than sequencing the entire genome.
How does targeted sequencing work?
The first step in targeted sequencing involves capturing or isolating the desired sequences from a sample. This can be done using various methods, such as hybridization capture or amplicon-based approaches.
Hybridization capture
This method synthesizes first the short DNA or RNA probes that are complementary to the target sequences, and then these probes will be attached to a solid support like magnetic beads. The sample containing the DNA or RNA of interest is then added to the probe-bound beads, and the target sequences are captured or hybridized with the probes.
Amplicon-based approaches
Amplicon-based targeted sequencing involves amplifying specific regions of DNA using PCR (polymerase chain reaction) before sequencing. This method allows for more precise targeting of specific regions and can be used in combination with other techniques like hybridization capture.
Advantages of targeted sequencing
The following are the advantages of targeted sequencing:
- Cost-effective: As only specific regions are sequenced, targeted sequencing is a cost-effective alternative to whole-genome sequencing.
- Higher coverage: By focusing on specific regions, the depth of coverage can be increased, providing more accurate and reliable results.
- Less computational burden: As compared to whole-genome sequencing, using targeted sequences will generate less amount of data, which in turn will reduce the computational burden thereby allowing a faster data analysis.
- Suitable for low-quality samples: Targeted sequencing can be used on degraded or low-quality DNA samples, making it a valuable tool in forensic and ancient DNA studies.
Disadvantages of Targeted Sequencing
While targeted sequencing presents numerous benefits, it is equally important to acknowledge the limitations of this technique.
- Limited scope: Since targeted sequencing focuses only on specific regions of the genome, any genetic variations outside of these regions will be missed. This limitation might exclude potentially significant genomic data.
- Design complexity: The design of probes or primers for capturing specific sequences can be complex and time-consuming, particularly for large genomic regions or numerous targets.
- Bias in amplification: Amplicon-based approaches can introduce bias due to the differential amplification of target sequences. This bias can affect the accuracy of variant detection.
- Difficulty in detecting structural variants: Targeted sequencing is less effective in identifying structural variants such as large deletions, duplications, or rearrangements, as these often occur outside the targeted regions.
- Need for prior knowledge: To design an effective targeted sequencing experiment, researchers must have prior knowledge of the regions of interest. This requirement might limit the use of targeted sequencing in exploratory studies where the regions of interest are unknown.
Despite these limitations, targeted sequencing remains a valuable tool in genomic research due to its cost-effectiveness, higher coverage of target regions, and suitability for various applications. Understanding both the strengths and weaknesses of targeted sequencing can guide researchers in choosing the most appropriate sequencing method for their specific research objectives.
Applications of targeted sequencing
Targeted sequencing has numerous applications in various fields of research. Some examples include:
- Cancer research: By targeting specific genes or mutations known to be associated with cancer, targeted sequencing can aid in the identification and characterization of potential biomarkers.
- Infectious disease diagnosis: Targeted sequencing has been used to detect and characterize pathogens in clinical samples, helping in the diagnosis and treatment of infectious diseases.
- Evolutionary studies: By providing insights into the evolutionary relationships and genetic diversity, targeted sequencing can be used to study specific regions of genomes in different species.
- Personalized medicine: By sequencing only the relevant genes or mutations, targeted sequencing can assist in identifying potential therapeutic targets for individual patients based on their unique genetic profiles.
Considerations to Achieve a Successful Targeted Sequencing
Implementing careful planning and execution is required to ensure accurate and reliable results, and the following are some additional considerations for successful targeted sequencing:
Choosing the right method
In conducting targeted sequencing, there are various methods that are available and each of these methods has its own advantages and limitations, thus, you must carefully consider your research goals and sample type before deciding on a particular approach.
Quality control measures
Targeted sequencing relies on high-quality DNA or RNA samples for accurate results. Implement quality control measures to ensure the integrity of your samples, such as DNA quantification and assessment of sample purity.
Primer design
If using an amplicon-based approach, proper primer design is crucial for successful targeted sequencing, so take into account factors such as primer specificity, melting temperature, and potential secondary structures in your primers.
Library preparation
Whether using hybridization capture or amplicon-based approaches, library preparation is a critical step in targeted sequencing. Follow established protocols and use high-quality reagents to ensure efficient capture of the target sequences.
Data analysis
Since targeted sequencing generates large amounts of data that require careful analysis, utilizing a bioinformatics tool and software is recommended since it is specifically designed for targeted sequencing data analysis to extract meaningful insights from your results.
By considering these additional factors, you can increase the chances of success in your targeted sequencing experiments and maximize the potential of this powerful technique in your research.
Conclusion
Targeted sequencing is a powerful tool that allows researchers to study specific regions of DNA or RNA with high accuracy and cost-effectiveness. With its wide range of applications, this technique continues to contribute significantly to various fields of research and has the potential to drive further advancements in the future. So, we can conclude that targeted sequencing is a valuable addition to the genetic toolbox for studying and understanding complex biological systems. Overall, targeted sequencing offers significant advantages over whole-genome sequencing and will likely continue to be a crucial method in many areas of research. As technology continues to advance, targeted sequencing methods will only become more precise and efficient, making it an essential technique for researchers in the years to come.
So, if you are interested in focusing on specific regions of DNA or RNA in your research, consider using targeted sequencing as a cost-effective and reliable option. Keep exploring and learning about the various methods and applications of targeted sequencing to stay at the forefront of genetic research.